I am working on a branching strategy in Github Enterprise, using mikepenz/release-changelog-builder-action to generate changelogs when new main versions are created. Changes are merged into a develop branch first, and when the action generates a changelog, it includes all the tags from the develop branch. All we want is to see the changes themselves, grouped by category (feature, fix etc.). Instead we get what you see in the picture. This is a test repo where I pushed one 'bug fix' but as you can see it is very cluttered with unnecessary tags. I have tried to search on Google as well as the action documentation, but either I don't understand it properly or it's not possible. Any help appreciated. This is the default, without a config file. Essentially, I want to remove everything boxed in red.
I figured out a workaround for this. The step that generates the changelog saves it as a .md file. So, I wrote a step that uses bash to take out what I don't want before the file is used to create a release. the lines I don't want start with either a single # with a space after it, ###, or -. Hopefully this is useful to someone.
- name: Remove clutter from changelog
id: trim-changelog
run: |
cat release_changelog.md | grep -e '^# ' -e ^### -e ^- > trimmed_changelog.md
Is there any way to render LaTex in README.md in a GitHub repository? I've googled it and searched on stack overflow but none of the related answers seems feasible.
For short expresions and not so fancy math you could use the inline HTML to get your latex rendered math on codecogs and then embed the resulting image. Here an example:
- <img src="https://latex.codecogs.com/gif.latex?O_t=\text { Onset event at time bin } t " />
- <img src="https://latex.codecogs.com/gif.latex?s=\text { sensor reading } " />
- <img src="https://latex.codecogs.com/gif.latex?P(s | O_t )=\text { Probability of a sensor reading value when sleep onset is observed at a time bin } t " />
Which should result in something like the next
Update: This works great in eclipse but not in github unfortunately. The only work around is the next:
Take your latex equation and go to http://www.codecogs.com/latex/eqneditor.php, at the bottom of the area where your equation appears displayed there is a tiny dropdown menu, pick URL encoded and then paste that in your github markdown in the next way:
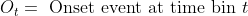
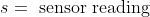
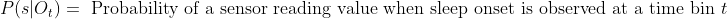
I upload repositories with equations to Gitlab because it has native support for LaTeX in .md files:
```math
SE = \frac{\sigma}{\sqrt{n}}
```
The syntax for inline latex is $`\sqrt{2}`$.
Gitlab renders equations with JavaScript in the browser instead of showing images, which improves the quality of equations.
More info here.
Let's hope Github will implement this as well in the future.
My trick is to use the Jupyter Notebook.
GitHub has built-in support for rendering .ipynb files. You can write inline and display LaTeX code in the notebook and GitHub will render it for you.
Here's a sample notebook file: https://gist.github.com/cyhsutw/d5983d166fb70ff651f027b2aa56ee4e
Readme2Tex
I've been working on a script that automates most of the cruft out of getting LaTeX typeset nicely into Github-flavored markdown: https://github.com/leegao/readme2tex
There are a few challenges with rendering LaTeX for Github. First, Github-flavored markdown strips most tags and most attributes. This means no Javascript based libraries (like Mathjax) nor any CSS styling.
The natural solution then seems to be to embed images of precompiled equations. However, you'll soon realize that LaTeX does more than just turning dollar-sign enclosed formulas into images.
Simply embedding images from online compilers gives this really unnatural look to your document. In fact, I would argue that it's even more readable in your everyday x^2 mathematical slang than jumpy .
I believe that making sure that your documents are typeset in a natural and readable way is important. This is why I wrote a script that, beyond compiling formulas into images, also ensures that the resulting image is properly fitted and aligned to the rest of the text.
For example, here is an excerpt from a .md file regarding some enumerative properties of regular expressions typeset using readme2tex:
As you might expect, the set of equations at the top is specified by just starting the corresponding align* environment
**Theorem**: The translation $[\![e]\!]$ given by
\begin{align*}
...
\end{align*}
...
Notice that while inline equations ($...$) run with the text, display equations (those that are delimited by \begin{ENV}...\end{ENV} or $$...$$) are centered. This makes it easy for people who are already accustomed to LaTeX to keep being productive.
If this sounds like something that could help, make sure to check it out. https://github.com/leegao/readme2tex
Since May 2022, this has been officially supported:
Inline:
Where $x = 0$, evaluate $x + 1$
Blocks:
Where
$$x = 0$$
Evaluate
$$x + 1$$
One can also use this online editor: https://www.codecogs.com/latex/eqneditor.php which generates SVG files on the fly. You can put a link in your document like this:
 which results in:
.
I test some solution proposed by others and I would like to recommend TeXify created and proposed in comment by agurodriguez and further described by Tom Hale - I would like develop his answer and give some reason why this is very good solution:
TeXify is wrapper of Readme2Tex (mention in Lee answer). To use Readme2Tex you must install a lot of software in your local machine (python, latex, ...) - but TeXify is github plugin so you don't need to install anything in your local machine - you only need to online installation that plugin in you github account by pressing one button and choose repositories for which TeXify will have read/write access to parse your tex formulas and generate pictures.
When in your repository you create or update *.tex.md file, the TeXify will detect changes and generate *.md file where latex formulas will be exchanged by its pictures saved in tex directory in your repo. So if you create README.tex.md file then TeXify will generate README.md with pictures instead tex formulas. So parsing tex formulas and generate documentation is done automagically on each commit&push :)
Because all your formulas are changed into pictures in tex directory and README.md file use links to that pictures, you can even uninstall TeXify and all your old documentation will still works :). The tex directory and *.tex.md files will stay on repository so you have access to your original latex formulas and pictures (you can also safely store in tex directory your other documentation pictures "made by hand" - TeXify will not touch them).
You can use equations latex syntax directly in README.tex.md file (without loosing .md markdown syntax) which is very handy. Julii in his answer proposed to use special links (with formulas) to external service e.g . http://latex.codecogs.com/gif.latex?s%3D%5Ctext%20%7B%20sensor%20reading%20%7D which is good however has some drawbacks: the formulas in links are not easy (handy) to read and update, and if there will be some problem with that third-party service your old documentation will stop work... In TeXify your old documentation will works always even if you uninstall that plugin (because all your pictures generated from latex formulas are stay in repo in tex directory).
The Yuchao Jiang in his answer, proposed to use Jupyter Notebook which is also nice however have som drawbacks: you cannot use formulas directly in README.md file, you need to make link there to other file *.ipynb in your repo which contains latex (MathJax) formulas. The file *.ipynb format is JSON which is not handy to maintain (e.g. Gist don't show detailed error with line number in *.ipynb file when you forgot to put comma in proper place...).
Here is link to some of my repo where I use TeXify for which documentation was generated from README.tex.md file.
Update
Today 2020.12.13 I realised that TeXify plugin stop working - even after reinstallation :(
For automatic conversion upon push to GitHub, take a look at the TeXify app:
GitHub App that looks in your pushes for files with extension *.tex.md and renders it's TeX expressions as SVG images
How it works (from the source repository):
Whenever you push TeXify will run and seach for *.tex.md files in your last commit. For each one of those it'll run readme2tex which will take LaTeX expressions enclosed between dollar signs, convert it to plain SVG images, and then save the output into a .md extension file (That means that a file named README.tex.md will be processed and the output will be saved as README.md). After that, the output file and the new SVG images are then commited and pushed back to your repo.
I just published a new version of xhub, a browser extension that renders LaTeX (and other things) in GitHub pages.
Cons:
You have to install the extension once.
Pros:
No need to set up anything.
Just write Markdown with math
Display math:
```math
e^{i\pi} + 1 = 0
```
and line math $`a^2 + b^2 = c^2`$.
(Syntax like on GitLab.)
Works on light and dark background. (Math has text-color)
You can copy-and-paste the math just like text
As an example, check out this GitHub README:
You can get a continuous integration service (e.g. Travis CI) to render LaTeX and commit results to github. CI will deploy a "cloud" worker after each new commit. The worker compiles your document into pdf and either cuses ImageMagick to convert it to an image or uses PanDoc to attempt LaTeX->HTML conversion where success may vary depending on your document. Worker then commits image or html to your repository from where it can be shown in your readme.
Sample TravisCi config that builds a PDF, converts it to a PNG and commits it to a static location in your repo is pasted below. You would need to add a line that fetches pdfconverts PDF to an image
sudo: required
dist: trusty
os: linux
language: generic
services: docker
env:
global:
- GIT_NAME: Travis CI
- GIT_EMAIL: builds#travis-ci.org
- TRAVIS_REPO_SLUG: your-github-username/your-repo
- GIT_BRANCH: master
# I recommend storing your GitHub Access token as a secret key in a Travis CI environment variable, for example $GH_TOKEN.
- secure: ${GH_TOKEN}
script:
- wget https://raw.githubusercontent.com/blang/latex-docker/master/latexdockercmd.sh
- chmod +x latexdockercmd.sh
- "./latexdockercmd.sh latexmk -cd -f -interaction=batchmode -pdf yourdocument.tex -outdir=$TRAVIS_BUILD_DIR/"
- cd $TRAVIS_BUILD_DIR
- convert -density 300 -quality 90 yourdocument.pdf yourdocument.png
- git checkout --orphan $TRAVIS_BRANCH-pdf
- git rm -rf .
- git add -f yourdoc*.png
- git -c user.name='travis' -c user.email='travis' commit -m "updated PDF"
# note we are again using GitHub access key stored in the CI environment variable
- git push -q -f https://your-github-username:$GH_TOKEN#github.com/$TRAVIS_REPO_SLUG $TRAVIS_BRANCH-pdf
notifications:
email: false
This Travis Ci configuration launches a Ubuntu worker downloads a latex docker image, compiles your document to pdf and commits it to a branch called branchanme-pdf.
For more examples see this github repo and its accompanying sx discussion, PanDoc example,
https://dfm.io/posts/travis-latex/, and this post on Medium.
I have been looking around and found that this answer in another question works best for me. i.e. use githubcontent math renderer, e.g. to display:
Use this link
Beware of the latex needs to be url encoded, but otherwise work quite well for me.
If you are having issues with https://www.codecogs.com/latex/eqneditor.php, I found that https://alexanderrodin.com/github-latex-markdown/ worked for me. It generates the Markdown code you need, so you just cut and paste it into your README.md document.
You may also take a look on my tool latexMarkdown2Markdown which convert LaTeX to SVG and generate a table of content with chapter numbering.
Good news!
According to this blogpost, now GitHub supports Mathjax in readme files.
You can use in-line LaTeX inspired syntax using $ delimiters, or in-blocks using $$ delimiters.
Writing inline expressions:
This sentence uses $ delimiters to show math inline:
$\sqrt{3x-1}+(1+x)^2$
Writing expressions as blocks:
The Cauchy-Schwarz Inequality
$$\left( \sum_{k=1}^n a_k b_k \right)^2 \leq \left( \sum_{k=1}^n a_k^2
\right) \left( \sum_{k=1}^n b_k^2 \right)$$
Source: https://docs.github.com/en/get-started/writing-on-github/working-with-advanced-formatting/writing-mathematical-expressions
You can use markdowns, e.g.
=\beta_0&space;+&space;\beta_1&space;x&space;+&space;u)
Code can be typed here: https://www.codecogs.com/latex/eqneditor.php.
Edit: As germanium pointed out, it does not work for README.md but other git pages though no explanation is available.
My quick solution is this
step 1. Add latex to your .md file
$$x=\sqrt{2}$$
Note: math eqns must be in $$...$$ or \\(... \\).
step 2. Add the following to your scripts.html or theme file (append this code at the end)
<script type="text/javascript" async
src="https://cdn.mathjax.org/mathjax/latest/MathJax.js?config=TeX-MML-AM_CHTML">
Done!. See your eq. by loading the page.
In TCL source tree, unix/Makefile, I notice that header files are not part of dependency of any C source file. For example:
tclCmdAH.o: $(GENERIC_DIR)/tclCmdAH.c
$(CC) -c $(CC_SWITCHES) $(GENERIC_DIR)/tclCmdAH.c
This ends up, if I update tcl.h and run make, the change is not picked up at all. I need to totally clean the directory and then run make.
Did I miss anything? Or any reason is it done this way?
I'm trying to get proper html output from gcovr when source file is located relative from my root directory.
For example (I will mention two cases where gcovr works, and where it has problems):
CASE:1 - gcovr works without problems
My root directory is structured as follow,after I run gcovr from root with --html --html-details
source/myfile.c
obj/myfile.o, myfile.gcda, myfile.gcno, myfile.c.gcov
gcovr_reports/report.html, report_myfile.html
So everything is ok, and I have the html general report(report.html) as well as the detailed report (report_myfile.html).
CASE:2 - gcovr is not working properly
My root directory is structured as follow,after I run gcovr from root with --html --html-details)
../../../Common/Source/myfile.c
obj/Common/Source/myfile.o, myfile.gcda,myfile.gcno,^#^#^#Common#Source#gcovmyfile.gcov
gcovr_reports/report.html, report.C
Now as you can see, gcovr generates the "report.C" file within the gcovr_report/ directory
Also the general html report (report.html) with the summary is created, but not the detailed one of my source file "myfile.c" .
When I look into the obj directory it creates the following file (as you can see below):
^#^#^#Project#Common#Source#myfile.c.gcov
When I take a look into
^#^#^#Project#Common#Source#myfile.c.gcov,
the path is resolved as follow:
Source:../../../Project/Common/Source/myfile.c
but it should be:
Source:../../../../../../../Project/Common/Source/myfile.c
The gcovr command is:
C:\Python34\Scripts\gcovr -v -b -r C:\Project\UnitTests\myModule\module1 -- object-directory C:\Project\UTests\myModule\module1\test-obj\Common\Source -- html --html-details -o govr_report\report.html
Any idea what I'm doing wrong?
Gcovr filters the coverage data to only show files within your project, as determined by the -r root or any --filters.
Project: C:\Project\UnitTests\myModule\module1
File: ..\..\..\Common\Source\myfile.c
The file is not within the project, so it is excluded. This is usually what you want, because the coverage of any libraries you use tends to be irrelevant.
When this is wrong, you can take over filtering and define your own --filters. You can define multiple filters, and any one must match. To disable filters entirely, use an empty filter --filter "".
Filters are regexes that are matched against the absolute source file path.
Unfortunately, filters are currently broken for Windows. It is not possible to write filters that match multiple directories, unless your working directory is a parent directory of all target directories. For example, you could go to C:\ and use the following filters:
--filter Project\\UnitTests\\myModule\\module1\\
--filter Common\\Source\\
This will change in a future version of gcovr, so please try to avoid filters that contain backslashes as path separators.
Update: Since gcovr version 4, all filters MUST use forward slashes for path separators, even on Windows. See Using Filters in the Gcovr User Guide for details.
I'm working on some code with a partner. Our make files differ slightly courtesy of different build setups. Because of this, so far we have not been tracking this file. However it would be nice to have at least one of ours tracked. The problem is, when that is done and the other person runs hg update, their copy gets update and the code won't compile.
Is there a way to track the file, but have it such that you can update the working directory selectively? Or is there some other way I should deal with this problem?
This is a slight variant of the standard "how do I deal with a config file" question. The standard answer in SVN, Mercurial, and Git is: don't track the file, instead track <file>.example. Then each user copies that over to <file> and tweaks it as needed.
But Makefiles are a bit smarter than config files: they execute code and can include other files. In which case, it starts making sense to track the Makefile normally and have it include another local file if it's present that overrides the default rules. For instance, the following will work with GNU Make:
# pull in any local user tweaks
-include Makefile.local
MQ extension is the best and The Right Way (tm) to do it (not easiest, but...)
Store common part of file in repo, individual personalisation - in own MQ-patches
Is it possible to combine your Makefiles? Then there is not chance of losing your different configurations by not storing them in version control.
For example, you could add a conditional statement based on the username. My username is ryan and this code echos my name, but if it is run on your computer, it probably will echo "not ryan."
all:
if [ `whoami` = "ryan" ]; then echo "ryan"; else echo "not ryan"; fi